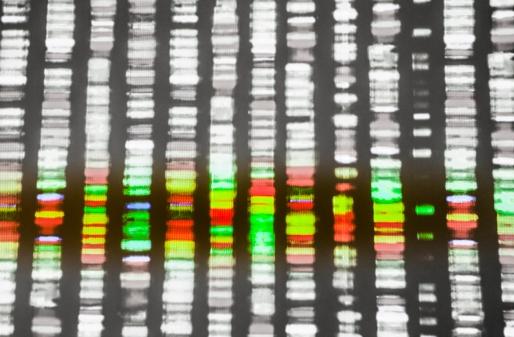
In the most detailed picture yet of Ebola's spread in an outbreak country, a gene sequencing study today revealed how the disease unfolded in Liberia, with most cases arising from a single introduction, sprouting multiple transmission chains within the country, then triggering more illness waves in Guinea.
West Africa's long and massive outbreak has yielded an unprecedented number of Ebola virus samples that researchers are pouring through to learn clues about how it spreads in regions and evolves in humans, findings that will help responders fine-tune their control efforts. After homing in on Liberia and comparing genetic patterns with neighboring countries, a team led by the US Army Medical Research Institute of Infectious Diseases (USAMRIID) published its findings in Cell Host & Microbe.
Gustavo Palacios, PhD, one of the senior authors, said in a USAMRIID press release, "The scope of this study has allowed us to piece together a complete picture of the Liberian portion of the outbreak. It also allowed us to investigate potential patterns of adaptation to the human host."
Liberia was the second worst-affected country and is 6 days into a 42-day countdown toward being declared free of the virus for a third time. The World Health Organization (WHO) said today in its weekly epidemiologic report that no new cases have been reported in the outbreak region and that investigations into a recent family cluster in Liberia are probably the result of rare reemergence of persistent virus from a survivor.
Comprehensive view of transmission
The USAMRIID team analyzed Ebola virus genomes from 139 patients infected during Liberia's second wave of infections, which was its largest. They compared them with 782 previously published sequences from throughout the outbreak, including those from Guinea, Sierra Leone, and Mali. Taken together, the sequences cover nearly 1 year of the epidemic.
Though they found evidence of many early introductions of the virus from Guinea and Sierra Leone, most of Liberia's cases were triggered by a single event in late May or early June of 2014, when Liberia's second wave began. Contact tracing shows a similar pattern, with three potential introductions from Sierra Leone during the same period, one of which led to several cases in Monrovia and went on to strike healthcare workers at Redemption Hospital.
The remote Lofa County was the likely point at which the Ebola virus entered Liberia. The team's analysis, however, suggests that two densely populated neighboring counties—Montserrado and Margibi—were the main sources for the spread of Ebola to Liberia's other counties.
That surge of cases not only led to the rapid spread and diversification of the virus within Liberia, but also seeded ongoing waves of the disease in Guinea and the exportation of the virus to Mali, according to the study.
Jason Ladner, PhD, first author of the study, said in a Cell Press news release, "The widespread movement of the Ebola virus within Liberia, due to a high rate of migration in the country, is likely to have played an important role in the magnitude and longevity of the Liberian portion of the Ebola outbreak." He added that regular migration of infected patients can complicate the surveillance and isolation steps needed to control the outbreak.
The team noted that demographic changes and internal conflict in West Africa have led to high migration rates in the region. Liberian census data from 2008 have suggested that 54% of Liberians over age 14 were internally displaced, and other sources have shown that 22% of Liberian-born people lived in other countries, which the researchers says have led to high migration rates and strong social ties across regions, especially in a relatively small country where regular travel is possible.
A detailed investigation of Ebola control measures used in West Africa, when combined with patterns revealed by gene studies, will help gauge the effectiveness of different management approaches, the authors said.
Little sign of increased adaptation
The outbreak's long span raised concerns in the global health community that continuing transmission might give the virus ample opportunities to evolve to adapt better to humans.
When the USAMRIID investigators looked at how the Ebola virus evolved, they identified diversification within Liberia, but no signs that it had further adapted to infected humans. "As a whole, we are seeing little evidence for additional adaptation of the virus to humans following the initial transition," they said in the study.
The substitutions they found were generally rare among the sequences they looked at, hinting that they probably didn't contribute much to the magnitude of West Africa's outbreak. And they noted that the findings were consistent with earlier virus evolution findings from Sierra Leone.
They added, though, that more research is needed to flesh out how the virus infected humans at the start of the outbreak. Targeted investigations into the earliest substitutions will be critical for understanding the transition of the virus to humans, the group wrote.
Similar studies planned for Guinea, Sierra Leone
Though the team looked at sequences from the other outbreak countries, their primary goal was identifying patterns and possible virus changes in Liberia.
Ladner said in the USAMRIID press release that the team's next steps are to look at patterns specific to Guinea and Sierra Leone.
"Over the next few months we will be combining forces with other research groups to look carefully at the commonalities and differences of Ebola virus spread and diversification within and between these three countries," he said. "This analysis should help to shed light on the most successful approaches for controlling the spread of the virus."
Palacios added that another step will be to functionally explore the different genetic mutations that have occurred during the outbreak. He said while computational analysis can identify possible functional changes, they need to be tested in experiments to see if the mutations affected virus fitness and pathogenicity.
"In particular, it will be very interesting to examine several of the changes that occurred early during the West African outbreak," he said.
See also:
Dec 9 Cell Host Microbe study
Dec 9 USAMRIID press release
Dec 9 Cell Press news release
Dec 9 WHO weekly Ebola update