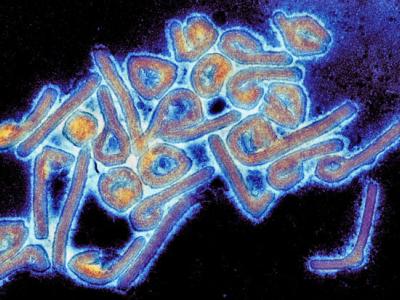
The Centers for Disease Control and Prevention (CDC) reported today that public health laboratories identified more than 220 samples of "nightmare" bacteria containing unusual resistance genes in 2017, a finding that officials say illustrates the importance of the agency's efforts to identify emerging drug-resistant pathogens quickly and contain them before they can spread.
In a new Vital Signs report, CDC researchers said that overall, more than 1,400 isolates of carbapenemase-producing bacteria were identified from clinical samples from 32 states during the first 9 months of 2017. Carbapenemase genes confer resistance to carbapenems, a class of powerful antibiotics that are considered a last resort for drug-resistant bacterial infections, and can be spread easily among different types of bacteria.
Carbapenem-resistant infections are exceedingly difficult to treat and have a mortality rate of nearly 50%.
While most of the isolates were carrying a carbapenem resistance gene that is familiar in the US healthcare system, 221 isolates were carrying resistance genes that are less common in the United States but are known to cause and spread carbapenem-resistant infections and outbreaks in hospitals in other parts of the world.
In addition, screening of healthcare contacts of infected patients by seven regional public health labs found that 1 in 10 were colonized with these bacteria.
CDC Principal Deputy Director Anne Schuchat, MD, said at a news conference that while the findings are troubling given the severity of carbapenem-resistant infections and their ability to spread in healthcare settings, they highlight some good news: The agency's updated containment strategy for new and emerging forms of antibiotic resistance appears to be having an impact.
"The bottom line is that resistance genes with the capacity to turn regular germs into nightmare bacteria have been introduced into many states, but with an aggressive response, we've been able to stomp them out promptly, and stop their spread between people, between facilities, and between other germs," Schuchat said.
Direct response helps slow CRE spread
The findings are part of a larger surveillance study that also analyzed infection data from the National Healthcare Safety Network (NHSN) over a 10-year period, from 2006 through 2015. The investigators were looking at these data to determine the percentage of Enterobacteriaceae carrying carbapenemase or extended-spectrum beta-lactamase (ESBL) genes that were reported to NHSN by US healthcare facilities, including acute care hospitals and nursing homes. ESBLs, which confer resistance to beta-lactam antibiotics, can also limit treatment options for infections.
ESBL-producing Enterobacteriaceae were first reported in the United States in 1988, and carbapenem-resistant Enterobacteriaceae (CRE) began to appear in US hospitals in 2001. But while healthcare facilities generally responded to the emergence of ESBL-producing isolates with their own infection control strategies, the approach to CRE, led by the CDC, has been more aggressive. In 2009, the CDC created CRE-specific guidance for healthcare facilities with recommendations for when CRE was detected, including lab surveillance of clinical cultures and targeted patient screening to identify healthcare contacts who might have acquired the bacteria but showed no signs of infection.
The investigators wanted see whether this more aggressive approach had resulted in differences in the spread of these two pathogens.
The data showed that among short-stay acute care hospitals, the percentage of Klebsiella pneumoniae and Escherichia coli isolates with an ESBL phenotype remained relatively stable, ranging from 17.6% (116 of 659 isolates) in 2006 to 16.5% (694 of 4,211) in 2015, and declined by 2% per year. But the proportion of isolates with a CRE phenotype fell by 15% per year, declining from 10.6% (64 of 604) in 2007 to 3.1% (115 of 3,718) in 2015.
"The difference seen may be due in part to the more directed response to slow the spread of the second germ, the nightmare bacteria CRE, once it was identified," Schuchat said.
The 2017 data, collected to evaluate the performance of an enhanced containment strategy introduced by the CDC in 2017, revealed that, out of 4,442 CRE and 1,334 carbapenem-resistant Pseudomonas aeruginosa (CRPA) isolates tested, 1,401 CRE (32%) and 25 CRPA (1.9%) were carbapenemase producers.
Most of those isolates expressed the K pneumoniae carbapenemase (KPC) gene, which is the most common carbapenemase gene detected in the United States. But 221 expressed four other carbapenemase genes—imipenemase (IMP), New Delhi metallo-beta-lactamase (NDM), Verona integrin encoded metallo-beta-lactamase (VIM), and oxacillinase-48-like carbapenemase (OXA-48).
To identify asymptomatically colonized healthcare contacts of infected patients, scientists conducted 1,489 screening tests in 50 facilities. Overall, 11% of the screening tests were positive for one of the five carbapenemases.
Enhanced containment
The agency's enhanced containment strategy focuses on quickly identifying unusual antibiotic resistance and resistance mechanisms in patients, isolating infected patients and assessing infection control in the healthcare facility where the resistance is found, screening for asymptomatic colonization, coordinating with other healthcare facilities, and continuing assessment and screening until the spread is controlled. It also calls for healthcare facilities and public health departments to respond to single isolates of emerging antibiotic-resistant pathogens like CRE.
The isolates collected in the study were identified by public health departments that are part of the CDC's Antibiotic Resistance Lab Network (ARLN), which was established in 2016 to improve the national capacity to rapidly detect and respond to antibiotic resistance. ARLN provides carbapenemase testing at 56 state and local public health departments and screening for asymptomatic carriage at 7 regional labs.
"The detection and response capacities from the newly established Antibiotic Resistance Lab Network, and stronger state-based antibiotic resistance response efforts, are improving prevention and response nationwide," Schuchat said.
One example of how this strategy has been effectively implemented comes from Iowa. In April 2017, rapid testing by the Iowa Department of Public Health (IDPH) and the CDC detected the IMP gene in Proteus bacteria isolated from a nursing home resident who had a urinary tract infection. Over the following weeks, nursing home staff and the IDPH assessed the facility for infection control gaps, followed contact precautions, and screened 30 residents. Five residents were found to carry the resistance gene, but the spread was stopped and no additional cases were found after several weeks.
"We'll see more successful cases like Iowa's as state and local public health leaders continue to champion this approach," Schuchat said, noting that not all clinicians and healthcare facilities have been aware of the help they can get from the CDC and state and regional public health departments.
"One of the messages that we want to send with this is that no provider has to go it alone," said Arjun Srinivasan, MD, associate director for healthcare-associated infection prevention programs at the CDC. "This is not something where you should feel like you've uncovered this situation and you don't have resources to help you handle it."
The authors of the paper estimate that for CRE alone, the containment strategy could reduce transmission in a single state by 20%, which would prevent 1,600 new infections over 3 years. The strategy can also be used for other emerging pathogens, including Candida auris and pan-resistant bacteria. The important thing, said Srinivasan, is that there has to be complete follow-through by everyone involved once a unique drug-resistant pathogen is detected.
"This is not a one-and-done thing," he said. "We keep at it…so the infection control team and the hospital, the providers, the health department, continue to look, continue to assess, and, if necessary, continue to test, until they know that the spread is controlled."
Health officials seeing an impact
Jay Butler, MD, chief medical officer for the Alaska Department of Health and Social Services, said the strategy, by providing better lab and epidemiologic response capacity, is paying benefits for providers who daily see the impact of drug-resistant infections.
"We're already seeing the impact of the CDC resources in improving our ability to prevent infections and our capacity to detect and respond to antimicrobial resistance," he said. "We can't wait until 1 case becomes 10, or 10 cases become 100; we can intervene early and aggressively to stop spread and to keep these threats out of our states."
Schuchat said the containment strategy for new and unusual drug-resistant threats complements the CDC's other efforts to combat antibiotic resistance, including strategies to prevent healthcare-associated infections and improve antibiotic use. But she warned that the agency needs to do more, and "we need to do it faster and earlier with each new antibiotic resistance threat."
"The hard truth is that as fast as we have run to slow resistance, some germs have outpaced us," she said. "We've had some success, but it just isn't enough to turn the tide."
See also:
Apr 3 CDC Vital Signs report
Apr 3 CDC additional Vital Signs material