Nigeria reports Lassa fever uptick as groups announce vaccine partnership
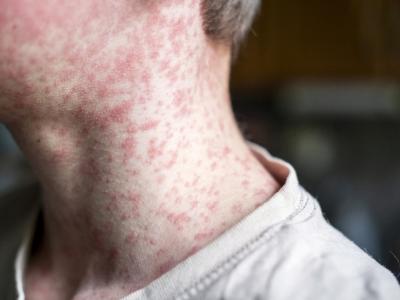
Lassa fever activity in Nigeria is increasing again, following an unprecedented outbreak earlier this year for which the acute phase was declared over on May 10, the World Health Organization's (WHO's) African regional office reported today in its weekly update.
The disease is endemic in the country, and though cases are reported in Nigeria year-round, illnesses typically peak between December and June. In West Africa, Lassa virus is spread by African soft-furred rats. People can become infected through contact with the rats' urine or feces, and the virus can spread from person to person through contact with body fluids.
In the week ending Dec 2, 13 confirmed cases, 3 of them fatal, were reported in five states: Ondo, Edo, Gombe, Kano, and Plateau. And over the past 5 weeks officials noted 32 cases, including 6 deaths, in six states, though most were in Edo and Ondo. Since Jan 1, Nigeria has reported 581 lab-confirmed cases, including 147 deaths. Over the course of the outbreak, 44 health worker infections have been reported, 3 of them in the past 4 weeks.
Dec 11 WHO African regional office report
In other Lassa fever developments, Batavia Biosciences, based in the Netherlands, today announced a partnership with the International AIDS Vaccine Initiative to develop a Lassa fever vaccine based on a vesicular stomatitis virus (VSV)-vectored system.
In a press release, the company said the goal is to develop a low-cost, easy-to-implement manufacturing system to speed the rapid clinical development of the vaccine and establish a stockpile.
Lassa virus is thought to cause an estimated 100,000 to 300,000 cases in Africa each year, about 5,000 of them fatal. Currently, no vaccine is available, and global health groups have prioritized the disease for vaccine development support and rapid review.
Dec 11 Batavia Bioscience press release
Global flu activity picks up; 91% of samples show influenza A
Though overall flu activity in the Northern Hemisphere remains low, the WHO said today in its latest global flu update that activity was on the rise, with influenza A representing more than 90% of detections.
Globally, of samples that tested positive for flu during the second half of November, 90.9% were influenza A and 9.1% were influenza B. Of the subtyped influenza A strains, 85.5% were 2009 H1N1, and 14.5% were H3N2.
Canada recorded an earlier-than-usual start to the flu season, with an uptick in pediatric hospitalization tied to H1N1, the WHO said. The US reported low activity but an increase in influenza-like illness (ILI).
Activity was low in Europe and Central Asia, with influenza A and B strains co-circulating. Korea recorded ILI above the seasonal threshold, while countries throughout western Asia reported low to no flu detections.
Throughout the Caribbean, Central America, and tropical South America, influenza detections were low, the WHO said.
Dec 11 WHO update
Report provides snapshot of November's MERS cases
In an overview of MERS-CoV cases reported for November, eight illnesses were reported, all in Saudi Arabia, the WHO Eastern Mediterranean regional office (EMRO) said today.
Two of the patients died from their MERS-CoV (Middle East respiratory syndrome coronavirus) infections, and two cases were secondary illnesses linked to household contact.
November's cases lift the global total since the virus was first detected in humans in 2012 to 2,274, at least 806 of them fatal. Saudi Arabia has reported most of the cases.
The WHO said the demographic and epidemiologic patterns of the disease haven't changed. But it noted that, since 2015, improved infection prevention and control practices have brought down the number hospital-acquired cases. The highest-risk group for primary infection is still adults ages 50 to 59, with those ages 30 to 39 at most risk for contracting secondary illnesses.
Dec 10 WHO EMRO update