In a study published last year in The Lancet, an international team of researchers estimated that 1.27 million people died in 2019 from an antibiotic-resistant infection, and nearly 5 million deaths were associated with antimicrobial resistance (AMR).
The study—one of the first to put hard numbers on the global burden of AMR—revealed that drug-resistant bacterial pathogens are a major global health threat and are as deadly as infectious diseases like HIV and malaria. And while they are threat to the whole world, the highest mortality burden is in low- and middle-income countries (LMICs), particularly those in sub-Saharan Africa and South Asia.
"High bacterial AMR burdens are a function of both the prevalence of resistance and the underlying frequency of critical infections such as lower respiratory infections, bloodstream infections, and intra-abdominal infections, which are higher in these regions," the authors of the study wrote.
The study, along with many others that have been conducted in LMICs, cited several reasons why AMR has a disproportionate impact in low-resources settings. Among them: inappropriate use of antibiotics that can be easily purchased without a prescription, counterfeit and substandard antibiotics, and poor sanitation and hygiene. All play a role in driving up resistance rates.
But perhaps the biggest factor is the lack of the diagnostic tools that can determine the right antibiotic needed for a patient's infection—or if an antibiotic is needed at all. The limited availability of these diagnostics in poor countries—whether it's an automated system that can identify the specific bacteria causing an infection and test for antibiotic susceptibility, or a rapid test that can determine whether an infection is bacterial or viral—is hampering these countries' ability to address the growing AMR threat at the most basic level.
"The availability of diagnostics aids in supporting or informing prudent use of antimicrobial medicines," Otridah Kapona, PhD, a laboratory scientist who specializes in AMR at the Zambia National Public Health Institute, told CIDRAP News. "And the opposite is true: Lack of diagnostic capacity, I would say, supports inappropriate use of antimicrobial medicines."
Lack of diagnostic capacity in hospitals, community
Kapona, who has been involved in the development and implementation of Zambia's first National Action Plan on AMR, says the lack of diagnostic capacity in countries like Zambia plays out at several levels.
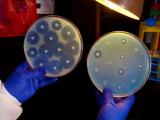
In most hospitals in wealthier countries, clinicians have access to expensive, automated systems or molecular methods that can provide quick pathogen identification and conduct antibiotic susceptibility testing (AST) directly from patient samples. But many hospitals in LMICs rely on traditional culture-based methods, in which bacteria from patient samples need to be grown, or cultured, to identify the specific pathogen and conduct antibiotic susceptibility testing—a process that can take 2 to 3 days.
As a result, clinicians in these settings often don't know the specific bacterium that's causing the infection and end up treating patients based on their symptoms and their own clinical experience. The typical result is treatment with broad-spectrum antibiotics that cover a wide range of bacteria but can promote resistance.
"We are targeting pretty much everything," Kapona said. "We are not specific in our management, and use of medicines like that accelerates the rate at which antimicrobial resistance develops in countries like Zambia."

Similar scenarios are seen in other LMICs that lack the type of sophisticated diagnostic systems that could enable clinicians to quickly determine the specific antibiotic needed. A 2020 study by researchers with the Center for Disease Dynamics, Economics & Policy found that use of "Watch" antibiotics—a label given to broad-spectrum drugs that the World Health Organization (WHO) says should not be used for routine infections because of their higher potential for promoting resistance—rose by 165% in LMICs from 2000 to 2015.
Limited diagnostic capabilities at hospitals and a lack of clinical reference labs that can perform testing for hospitals aren't the only reasons for that increase; poor sanitation and a higher incidence of drug-resistant infections are also factors. But the lack of diagnostic capability plays a significant role.
"There are very few labs that are able to do microbiology, where they can correctly identify an organism and do antimicrobial susceptibility testing to inform what antimicrobials the doctor should use to treat that particular patient or client," Kapona said.
And that has a cascading effect, according to Cecilia Ferreyra, MD, director of the AMR program at the nonprofit Foundation for Innovative New Diagnostics (FIND). If hospital clinicians aren't sending bacterial samples to be cultured and tested for susceptibility because the process takes too long, then hospitals don't have a sense of what resistance profiles are for various pathogens and can't develop accurate treatment guidelines.
"When a patient goes to a hospital and we don't have diagnostics that can quickly tell me whether that patient has a Klebsiella infection or a Staph infection…I won't know what to use," Ferreyra said. "And because there is this overall lack of data on what is a resistant profile in those settings, I will be prescribing something that might or might not be really useful at all."
Lack of diagnostics also affects antibiotic prescribing at the community level in low-resource settings, where small primary care clinics have even fewer resources. As Ferreyra, Kapona, and their colleagues noted in a paper published last year in PLOS Global Public Health, the most widely used test in those settings to determine whether an infection is viral or bacterial—C-reactive protein (CRP) tests—can't distinguish whether elevated CRP levels are caused by bacteria or by malaria, dengue, or COVID-19. And other tests on the market are too expensive for low-resource settings.
There are very few labs that are able to do microbiology, where they can correctly identify an organism and do antimicrobial susceptibility testing to inform what antimicrobials the doctor should use to treat that particular patient or client.
Without rapid, accurate, and affordable point-of-care tests that can quickly distinguish whether an infection is bacterial or viral, adults and children who walk in with a fever or respiratory symptoms caused by a virus are likely to walk out with antibiotics, which are often viewed as a quick and easy solution.
"When you are seeing ambulatory patients, patients coming in with cough, fever, diarrhea, or malaria, if you don't have an available point-of-care [test] to quickly identify what the cause of the fever is, the patients will walk back home probably having at least one antibiotic that they may or may not need," Ferreyra said.
And that may affect future healthcare-seeking behavior. If patients with a fever have to travel a long distance to a clinic and all they get is a quick assessment and an antibiotic, Kapona explains, the next time they're feeling feverish, they might skip the assessment and go right to the treatment.
"They'll just go to the first pharmacy or chemist and [get] the same medication," she said. "And unfortunately, because of weak regulatory systems, people can access these medications off the counter without a prescription."
Efforts to strengthen hospital diagnostics, surveillance
While limited access to diagnostics has not received the same type of attention that has been given to the weak pipeline of new antibiotics, it's hasn't gone unrecognized. That's in part because the WHO and other organizations working to address the AMR crisis realize that the ability of poor countries to conduct AMR surveillance at a national level requires greater diagnostic capacity.
One of the companies that's working to address the issue is French diagnostics company bioMérieux. In a partnership with UK-based Fleming Fund, which supports international AMR monitoring and surveillance efforts, bioMérieux is equipping clinical reference and veterinary labs in 15 countries across Africa (including Zambia) and the Asia-Pacific with automated mass-spectrometry systems that can identify bacterial pathogens directly from various clinical specimens and then conduct susceptibility testing in the AST platform, providing results in a matter of hours.

The company is also providing IT-integrated software tools, known as middleware, that assist microbiologists in interpreting results and providing them to clinicians.
"bioMérieux's mission is to integrate diagnostics solutions in reference labs and support antimicrobial stewardship by providing a comprehensive middleware solution," said Michel Bonnier, Global Health Senior Director at bioMérieux. "As a Fleming fund partner, we aim to prevent antimicrobial resistance by delivering accurate data and actionable insights to healthcare professionals."
The Fleming Fund's hope is that the data collected by the labs will not only help clinicians in these countries prescribe the right antibiotics at the right time to patients but also help hospitals develop treatment guidelines. In addition, the data will be sent to national surveillance labs and uploaded to the WHO Global Antimicrobial Surveillance and Use System (GLASS), which will inform global AMR surveillance efforts.
That's significant, because the current lack of capacity to collect and interpret data on resistant pathogens in LMICs means national AMR surveillance data are limited. In fact, the authors of the Lancet study noted that the limited AMR surveillance data in low-resource countries, and the fact that many lab-based surveillance systems in these countries aren't linked to patient outcomes, likely means the burden of death from resistant infections could be much higher than they estimated.
Bonnier says bioMérieux is not simply providing the equipment. It's also training lab workers in the countries to use the systems and providing maintenance and technical support. "We want this to a be a sustainable program," he said.
The training is an important component of building that sustainability. As Kapona explains, while the equipment for quicker pathogen identification and susceptibility testing is necessary, having trained staff who can correctly use it is and interpret the information is equally important.
"Even if you were to have the equipment available today…building this kind of human resource capacity takes time because it has to be a continuous hands-on training of staff," she said.
Smartphone solutions
There are also efforts to bring cheaper diagnostic solutions to low-resource settings. Among them is Antibiogo, a mobile app developed by the humanitarian medical group Medecins Sans Frontieres (MSF) that enables lab technicians without clinical microbiology training to interpret antibiotic susceptibility test results using their smart phones. The application has already been deployed in Yemen, the Democratic Republic of the Congo, the Central African Republic, Mali, and Yemen.
"With Antibiogo, any microbiology laboratory technician—anywhere in the world—will be able to read and interpret an AST directly on any phone and know the resistance profile of the bacteria responsible for patient infection," Nada Malou, PhD, head of the Antibiogo program, said in an MSF press release.
"What this app is doing is amazing," Ferreyra said.
Another smartphone-based test that could help low-resource clinics with pathogen identification is smaRT-LAMP, developed by scientists at the University of California, Santa Barbara. The system, which can be produced for less than $100, includes a hot plate, chemicals to amplify viral or bacterial DNA from patient samples, a cardboard box, and LED lights. A smartphone camera captures the fluorescent signal (illuminated by the LED lights) from the chemical reaction, and a custom-built app analyzes the image to make a diagnosis.
You need to have diagnostics at every point that you are seeing a patient to be able to prescribe in an informed way and preserve antibiotics.
The COVID-19 pandemic has opened up a world of possibilities for this device. In a study published in JAMA Network Open last year, the developers of the test showed that smART-LAMP matched the performance of polymerase chain reaction (PCR) tests in detecting SARS-CoV-2 and influenza in saliva samples. And in a 2018 study in EBioMedicine, smaRT-LAMP showed it could identify bacteria in urine samples as accurately as PCR tests—at a fraction of the cost and the time.
A device like smaRT-LAMP could help healthcare clinics in LMICs quickly identify urinary tract infections. But Michael Mahan, PhD, a professor at UC Santa Barbara who helped develop the system, says it also could be modified to determine the most appropriate antibiotic treatment.
"Primers could be designed for the detection of specific genes encoding antibiotic resistance," he said.
Ultimately, says Ferreyra, low-resource countries will need to have diagnostic capability at all levels—in community clinics, in hospital laboratories, and at the surveillance level—to be able to give patients the best treatment possible and help clinicians practice antibiotic stewardship.
"You need to have diagnostics at every point that you are seeing a patient to be able to prescribe in an informed way and preserve antibiotics," she said.