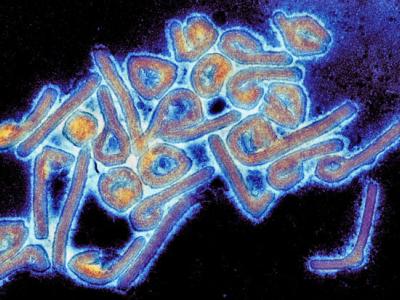
New research by a team of European scientists indicates that the rise of carbapenem-resistant Klebsiella pneumoniae in Europe is being fueled mainly by four strains that are being spread among hospital patients.
In a study yesterday in Nature Microbiology, researchers with the Wellcome Sanger Institute and partners from several European universities performed genome sequencing on a collection of more than 1,700 K pneumoniae isolates from patients in 244 hospitals across Europe. Their analysis revealed that four clonal lineages appear to be responsible for most of the carbapenem-resistant K pneumoniae isolates, and that roughly half of the isolates were closely related to another isolate collected at the same hospital.
The authors of the study say the findings highlights the fact that, while there has been an increasing focus on antibiotic use in animals and how the spread of resistance genes in the environment might affect human health, hospitals remain the primary source of transmission for multidrug-resistant pathogens.
Rising threat of carbapenem resistance
The isolates analyzed for the study came from the European Survey of Carbapenemase-Producing Enterobacteriaceae (EuSCAPE), a multinational survey of European hospitals in which carbapenem non-susceptible and susceptible K pneumoniae and Escherichia coli isolates were collected from 455 European hospitals over a 6-month period in 2013 and 2014. Data from that survey had previously been used to estimate the incidence of carbapenemase-producing Enterobacteriaceae in Europe's hospitals.
Carbapenems are one of the most powerful antibiotics used in hospitals to treat severe bacterial infections, and the rise of carbapenem resistance raises the prospect of infections that are nearly impossible to treat.
Over the past 15 years, the incidence of infections and the number of deaths linked to carbapenem-resistant K pneumonia, which can cause severe respiratory, urinary tract, and bloodstream infections, has been rising at an alarming rate in European hospitals. In a study published last year in the The Lancet Infectious Diseases, researchers estimated that the median number of carbapenem-resistant K pneumoniae infections rose from 2,535 in 2007 to 15,910 in 2015, while attributable deaths rose from 341 to 2,094.
For this study, the researchers wanted to know more about what the carbapenem-resistant K pneumoniae isolates collected in EuSCAPE looked like—what kind of resistance mechanisms they were carrying, where they were coming from, and which strains were causing infections. By analyzing the genome sequences of the K pneumoniae isolates from this collection, they were able to determine which carbapenemase genes they were carrying.
Carbapenemase genes carry enzymes that confer resistance to carbapenem antibiotics and are seen as crucial to the emergence and spread of carbapenem resistance in K pneumoniae and other pathogens. They are often carried on mobile pieces of DNA that can be shared within and between different species of bacteria.
Patient-to-patient, between-hospital spread
Of the 1,717 K pneumoniae isolates with genome sequences, 944 (55.0%) were carbapenem non-susceptible, and 69.6% of those isolates carried one or more of the five most frequently reported carbapenemase genes worldwide: blaKPC-like, blaOXA-48-like, blaNDM-like, blaVIM-like, and blaIMP-like. Nearly 70% of the isolates carrying these carbapenemase genes were grouped in one of four major clonal lineages of K pneumoniae that have recently emerged and have been the most successful worldwide: sequence types (STs) 11, 15, 101 and 258/512 (considered a single lineage).
"These lineages bear the hallmarks of so-called 'high-risk' clones, which commonly share a recent ancestor, epidemic success, and a defined geographic distribution," the authors of the study write. "They are also often associated with outbreaks, and probably possess particular characteristics that increase their tenacity, transmissibility and population size, providing a greater opportunity for the acquisition of antibiotic resistance genes."
Analysis of the genetic relatedness of the carbapenemase-carrying isolates revealed that in 52.6%, the genetically nearest neighbor (gNN) in the collection came from a patient in the same hospital. And of the 171 hospitals that contributed carbapenemase-carrying isolates, 96 (56.1%) had at least two isolates that were gNNs. Both of these findings are indications that patient-to-patient transmission is driving the spread of these carbapenem-resistant strains.
Carbapenemase-carrying K pneumoniae isolates from different hospitals in the same country were also more genetically similar than isolates from different countries, suggesting inter-hospital spread is occurring within countries.
Need for better infection prevention, control
The authors say the findings highlight the need for better infection prevention and control in hospitals and for genomic pathogen surveillance, which could enable hospitals to reduce the potential for transmission by identifying patients carrying these strains more quickly and isolating them from other patients.
"Genomic surveillance will be key to tackling the new breeds of antibiotic-resistant pathogen strains that this study has identified," paper co-author David Aanensen, PhD, director of the Centre for Genomic Pathogen Surveillance at the Wellcome Sanger Institute, said in a Wellcome press release.
"Currently, new strains are evolving almost as fast as we can sequence them. The goal to establish a robust network of genome sequencing hubs will allow healthcare systems to much more quickly track the spread of these bacteria and how they're evolving."
See also:
Jul 29 Nat Microbiol study
Jul 29 Wellcome Sanger Institute press release
Nov 5, 2018, Lancet Infect Dis study on deaths from resistant bacteria in Europe