A new study by scientists in Sweden indicates that bacteria exposed to small concentrations of antibiotics over time can become highly resistant, a finding the authors say provides an example of how low levels of antibiotics present in many environments may potentially contribute to antibiotic resistance.
In the study, published this week in Nature Communications, researchers from Uppsala University demonstrated that Salmonella exposed repeatedly to an amount of streptomycin that was not strong enough to kill the bacteria or inhibit growth still evolved high-level resistance. The resistance was caused by genetic mutations that haven't been typically associated with antibiotic resistance and were different from those that develop when the bacteria is exposed to lethal amounts of the drug.
"These results demonstrate how the strength of the selective pressure influences evolutionary trajectories and that even weak selective pressures can cause evolution of high-level resistance," the authors write.
Evolution of high-level resistance
To study the evolution of antibiotic resistance when exposed to low levels of antibiotics, the scientists repeatedly exposed a strain of streptomycin-susceptible Salmonella enterica serovar Typhimurium to an amount of streptomycin that was roughly one-quarter of the minimum inhibitory concentration (MIC)—the lowest concentration of an antibiotic needed to prevent bacterial growth. This amount was enough to reduce the bacterial growth rate by 3%. The bacteria were exposed to the streptomycin every 24 hours for 900 generations.
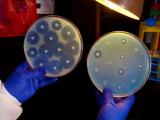
After this repeated exposure, the Salmonella Typhimurium bacteria were found to have varying levels of resistance, but four clones exhibited high levels of streptomycin resistance and were investigated further using whole-genome sequencing (WGS). This method of DNA fingerprinting revealed that the most resistant bacterial strains had acquired mutations in five genes, most of which are not typical resistance genes. In addition, the affected genes were different than the mutated gene found when the scientists exposed a Salmonella Typhimurium strain to an amount of streptomycin well above the MIC.
Individually, only one of these genetic mutations produced a significant increase in streptomycin resistance. But the scientists found that when the five mutant genes were combined they produced a much higher level of streptomycin resistance than expected, an indication that the resistance is being driven by the interaction of these mutations.
The findings are significant because they show that high-level antibiotic resistance doesn't only occur when bacteria are exposed to high levels of antibiotics; it can also evolve in response to low-level antibiotic exposure, driven by mutations in genes and that have not previously been associated with antibiotic resistance. This suggests that the number of genes associated with antibiotic resistance could be underestimated, and that these genes could act as a "stepping stone" for the evolution of clinically significant resistance.
Low levels of antibiotics in animals, environment
The suggestion that even small amounts of antibiotic exposure could be contributing to antibiotic resistance has big implications. Livestock and poultry producers routinely give their animals sub-therapeutic doses of antibiotics to prevent disease, a practice many experts believe contributes to antibiotic resistance.
In addition, a growing body of research conducted in recent years has documented the presence of antibiotics and antibiotic residues in rivers, lakes, tidal estuaries, and wastewater around the world, with households, hospitals, pharmaceutical factories, and farms as the likely sources.
According to a recent report from the United Nations Environment Programme, up to 80% of antibiotics used by humans are excreted un-metabolized through urine and feces into sewage systems and flow into wastewater treatment plants that also treat water from hospitals and industrial facilities. Because these plants aren't designed to fully remove antibiotics and other pharmaceuticals from wastewater, they end up being released into surface waters. Likewise, antibiotics used in food-animal production get excreted in manure and show up in farm soils or get carried into nearby lakes and streams with other untreated agricultural runoff.
Although the extent of environmental contamination from human and animal antibiotic use, and role that this contamination plays in the development of clinically relevant antibiotic resistance, are not yet clear, many experts believe the presence of even trace amounts of antibiotics in the environment is not without consequences.
Lead study author Dan Andersson, PhD, a professor in the department of medical biochemistry and microbiology at Uppsala, said the study provides evidence that the very low concentrations of antibiotics present in a number of environments could contribute to antibiotic resistance evolution. But how significant a contributor, in comparison to the selection pressure caused by antibiotic use in patients, is hard to say.
"It's likely to contribute, but it's hard to put a number on it," Andersson said in an email.
Andersson and his colleagues are planning similar studies with other antibiotics and other bacterial species, and hope to test their findings outside of the lab. "We want to examine if this selection process occurs outside of the laboratory, in natural settings," he said.
See also:
Apr 23 Nat Commun study